Overview
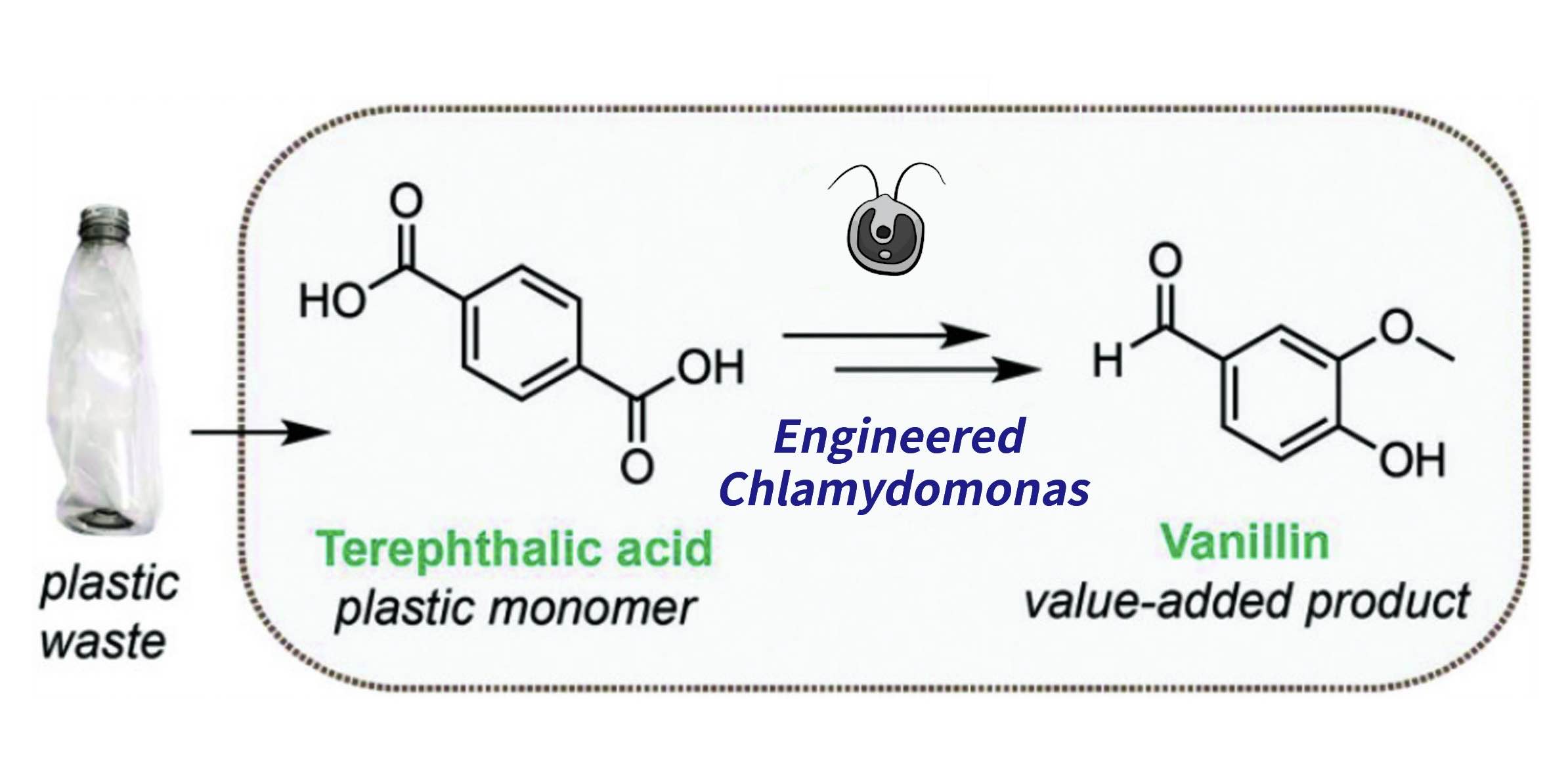
We decided to engineer Chlamydomonas reinhardtii with an enhanced enzyme system to upcycle plastic waste: This system will depolymerize Polyethylene terephthalate (PET) into reusable building blocks in order to upcycle PET into a high-end product, vanillin. Our goal is to address both the poor management of post-consumer plastic waste and the pressing need to apply circular economic solutions to those fossil fuel products.
The Problem
Imagine a world where our plastic waste transforms into valuable resources through synthetic biology. This vision is not only fascinating but essential, in a world where plastic production - and subsequently, though not to a proportional degree, waste - has more than doubled over the past two decades. Polyethylene terephthalate (PET), the most widely produced and common polymer worldwide, is most often found in bottles and packaging, contributing significantly to ocean and landfill pollution. Currently, about 70% of global plastics become waste. Only around 41% of post-consumer plastic waste is recovered through recycling or incineration, while 40% is disposed of in landfills and 19% makes its way into oceans or onto coastlines. At present, the accumulation of PET waste is steadily increasing and poses a growing threat to ecosystems worldwide. It is estimated that microorganisms in the environment take hundreds of years to fully degrade PET plastics. Developing microorganisms capable of degrading PET into vanillic acid holds promise as it transforms plastic waste into a valuable molecule used across various industrial sectors. This approach provides a sustainable pathway for utilizing plastic waste and supports a circular economy.
We all know how recycling is an important part of managing environmental pollution levels, but right now less than 20% of plastic waste goes through this process. We thus realized implementing upcycling as a key aspect of the project, so as not to simply degrade a compound but create a new high-value one, would enable us to organically implement it to a large-scale bioproduction. Such an approach would allow not only to degrade PET, but to valorise it. This led us to the search for a compound that would meet two requirements: to be feasibly synthesized through enzymatic processes, and be a highly sought out primary compound. We found that vanillin demands far exceed the offer, and got to work.
Our Solution
Recently, Japanese researchers identified the bacterium Ideonella sakaiensis, which is capable of producing the enzymes necessary to break down PET plastic into its two main components: TPA and EG. This transformation is made possible by the expression of two specific enzymes, named PETase and MHETase. The reactions of these enzymes are illustrated below.

This discovery has played a major role in the field of sustainable development, opening new perspectives for synthetic biology enthusiasts to create innovative solutions to the problem of microplastic pollution. Since then, numerous studies have been conducted and several discoveries have been made. Among these is the creation of a PETase-MHETase fusion protein with enhanced efficiency. Additionally, bacterial systems capable of converting PET into various molecules have been developed, allowing the degradation of PET and the production of economically valuable molecules from the degradation products, which has had a significant impact in the field of synthetic biology. In our project, we were inspired by this idea of transforming plastic into a more useful molecule. We aim to create a recombinant strain of Chlamydomonas reinhardtii capable of converting PET into vanillic acid (VA).
Why Chlamydomonas? Chlamydomonas reinhardtii is a well-studied, photosynthetic, unicellular eukaryote, making it a very interesting candidate for developing an autonomous photosynthetic system capable of converting PET into vanillic acid. Moreover, microalgae represent an attractive option for the synthesis of recombinant proteins, contrasting with traditional heterotrophic organisms such as E. coli, yeast, or CHO cells. Their low-cost phototrophic cultivation, as well as the generally recognized as safe (GRAS) status of multiple algae strains, including Chlamydomonas reinhardtii, prompted us to elect it as our main strain. Our work is divided into two main stages: The first stage involves introducing a gene encoding the PETase-MHETase fusion protein into the genome of Chlamydomonas reinhardtii to enable it to convert PET into TPA. To do this, we must first prepare the necessary plasmids using the MoClo technique, which allows the assembly of multiple basic parts simultaneously and the formation of the required plasmids (Crozet et al. 2018). The second stage aims to integrate the formed TPA into the metabolism of Chlamydomonas reinhardtii so that it serves as a carbon source, with the ultimate goal of producing vanillic acid from another metabolic molecule of Chlamydomonas
Creating our Plasmid
To achieve the first step of converting PET to TPA, we need to transform Chlamydomonas reinhardtii with the plasmid containing the gene for the PETase-MHETase fusion protein, which will enact the reaction PET -> MHET -> TPA.
The techniques and main steps of this process are inspired by the work of the 2020 iGEM team from Sorbonne University. To construct the plasmid, we use the MoClo technique, which allows the assembly of basic parts such as promoters, CDS, and terminators and which relies on the Golden Gate cloning strategy (Weber et al. 2011)
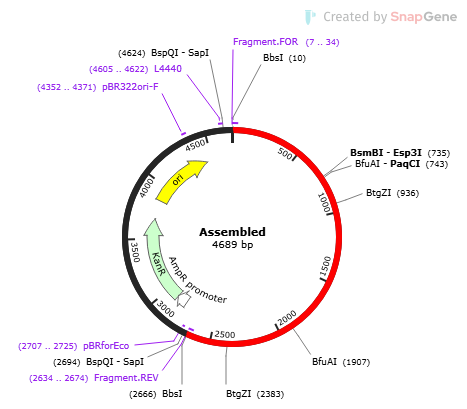
In the MoClo assembly standard (Weber et al. 2011), the smallest units are basic genetic parts such as promoters, CDS, 5’ UTRs, and tags. Each type has a specific position defined by the fusion sites on both sides. These basic units are cloned into plasmids using the BsaI restriction site and are referred to as "Level 0." A Level 0 acceptor plasmid (pAGM1311) contains BsaI restriction sites flanking a lacZ cassette, a pUC origin of replication, and the kanamycin resistance gene. Upon digestion with BsaI, the lacZ cassette is released, and the part of interest hybridizes to the plasmid through the complementarity of the flanking overhangs. These molecules are then ligated, and the final products are used to transform à strain of E. coli. Transformants carrying Level 0 plasmids are selected by blue/white screening All Level 0 plasmids also contain a SapI recognition site that allows a second golden gate reaction to create an entire transcription unit (Level 1) from several Level 0 plasmids (Weber et al. 2011).
What about CDS? In designing a coding sequence (CDS) for proteins of exogenous origin (in our case, of bacterial origin ) adapted to our Chlamydomonas reinhardtii , we need to reverse translate the protein sequence to generate cDNA. For this, we used the sequence of the fusion protein PETase-MHETase with a 12 amino acid linker and a 6 His Tag (Addgene plasmid pCJ190). The Chlamydomonas genome is very GC-rich, which implies that GC-rich codons are more frequently encountered. So it is necessary to adapt our sequences to the codon bias found in Chlamydomonas. The next step in designing our standardized sequence was to add the fusion sites corresponding to CDS and to add restriction sites for the type IIS restriction enzyme BsaI at the 5’ and 3’ ends. Finally, to avoid any internal hydrolysis of our gene parts during the digestion/ligation reaction, internal type IIS sites of BsaI and SapI were removed. Finally, the second step involves introducing TPA as a carbon source into the metabolism of Chlamydomonas and producing VA from another metabolic molecule of Chlamydomonas. At present, we are still searching for the best candidates, as the direct transformation of TPA into VA is very complicated to achieve in Chlamydomonas.
References
- Hannah Ritchie, Veronika Samborska and Max Roser (2023) - “Plastic Pollution” Published online at OurWorldInData.org. Retrieved from : 'https://ourworldindata.org/plastic-pollution' [Online Resource]
- Orlando, M.; Molla, G.; Castellani, P.; Pirillo, V.; Torretta, V.; Ferronato, N. Microbial Enzyme Biotechnology to Reach Plastic Waste Circularity: Current Status, Problems and Perspectives. Int. J. Mol. Sci. 2023, 24, 3877. https://doi.org/10.3390/ijms24043877
- Soong YV, Sobkowicz MJ, Xie D. Recent Advances in Biological Recycling of Polyethylene Terephthalate (PET) Plastic Wastes. Bioengineering (Basel). 2022 Feb 27;9(3):98. doi: 10.3390/bioengineering9030098. PMID: 35324787; PMCID: PMC8945055.
- Di Rocco, G., Taunt, H.N., Berto, M. et al. A PETase enzyme synthesised in the chloroplast of the microalga Chlamydomonas reinhardtii is active against post-consumer plastics. Sci Rep 13, 10028 (2023). https://doi.org/10.1038/s41598-023-37227-5
- Changko, S., Rajakumar, P.D., Young, R.E.B. et al. The phosphite oxidoreductase gene, ptxD as a bio-contained chloroplast marker and crop-protection tool for algal biotechnology using Chlamydomonas. Appl Microbiol Biotechnol 104, 675–686 (2020). https://doi.org/10.1007/s00253-019-10258-7
- Sadler JC, Wallace S. Microbial synthesis of vanillin from waste poly(ethylene terephthalate). Green Chem. 2021 Jun 10;23(13):4665-4672. doi: 10.1039/d1gc00931a. PMID: 34276250; PMCID: PMC8256426.
- https://static.igem.org/mediawiki/2019/8/89/T--Sorbonne_U_Paris--TheChlamyGuide.pdf
- https://2021.igem.org/Team:ASTWS-China/Design
- https://2020.igem.org/Team:Sorbonne_U_Paris/Description
- Benyathiar P, Kumar P, Carpenter G, Brace J, Mishra DK. Polyethylene Terephthalate (PET) Bottle-to-Bottle Recycling for the Beverage Industry: A Review. Polymers (Basel). 2022 Jun 11;14(12):2366. doi: 10.3390/polym14122366. PMID: 35745942; PMCID: PMC9231234
- Knott BC, Erickson E, Allen MD, Gado JE, Graham R, Kearns FL, Pardo I, Topuzlu E, Anderson JJ, Austin HP, Dominick G, Johnson CW, Rorrer NA, Szostkiewicz CJ, Copié V, Payne CM, Woodcock HL, Donohoe BS, Beckham GT, McGeehan JE. Characterization and engineering of a two-enzyme system for plastics depolymerization. Proc Natl Acad Sci U S A. 2020 Oct 13;117(41):25476-25485. doi: 10.1073/pnas.2006753117. Epub 2020 Sep 28. PMID: 32989159; PMCID: PMC7568301
- Tanasupawat S, Takehana T, Yoshida S, Hiraga K, Oda K. Ideonella sakaiensis sp. nov., isolated from a microbial consortium that degrades poly(ethylene terephthalate). Int J Syst Evol Microbiol. 2016 Aug;66(8):2813-2818. doi: 10.1099/ijsem.0.001058. Epub 2016 Apr 5. PMID: 27045688
- Biological Valorization of Poly(ethylene terephthalate) Monomers for Upcycling Waste PET Hee Taek Kim, Jae Kyun Kim, Hyun Gil Cha, Myung Jong Kang, Hyun Sook Lee, Tae Uk Khang, Eun Ju Yun, Dae-Hee Lee, Bong Keun Song, Si Jae Park, Jeong Chan Joo, and Kyoung Heon Kim ACS Sustainable Chemistry & Engineering 2019 7 (24), 19396-19406 DOI: 10.1021/acssuschemeng.9b03908



