Results
High-throughput Drug Screening, In Vitro Tumor Suppression Validation
Our high-throughput drug screening experiments were conducted in the laboratory, with the assistance of researchers and a Zhongxi Biological SC9210 fully automated pipetting workstation. Compounds for screening were obtained from the MCE compound library, specifically the MCE Novel Known Bioactive Compound Library (1,220 compounds). All compounds were dissolved in 100% DMSO or water. Prior to the experiment, the compounds were pre-diluted to prepare 1 mM stock solutions. The cells used for screening included MCF-10A normal mammary epithelial cells and MDA-MB-231 triple-negative breast cancer cells. The cells were seeded in 96-well culture plates, with a density of approximately 5,000 cells per well, in a total volume of 100 µL medium. After 24 hours of cell adhesion, 1 µL of each compound from the library was automatically added to the culture medium in the 96-well plates using the four-dimensional modular automation handling platform, resulting in a final drug concentration of 10 µM.
We set the negative control as the 10 µM pure DMSO group, and the positive control as the 10 µM cisplatin group. After 48 hours of drug treatment, the medium in the wells was discarded, ATP detection reagent was added, and luminance was measured using a microplate reader, or the effectiveness of ATP probes was assessed using a Revvity high-content analyzer.
During our high-throughput drug screening experiment, we conducted a standardized comprehensive screening (screening all 1,220 compounds). From this experiment, we identified 15 small-molecule compounds. These small molecules did not reduce the viability of normal mammary epithelial cells (MCF-10A) to below 80% (Above the red dotted line perpendicular to the Y-axis), while reducing the viability of breast cancer cells (MDA-MB-231) to below 30% (To the left of the red dotted line perpendicular to the X-axis).
The 15 small-molecule compounds are listed below:
- 2-(1-Aziridinyl)ethanol
- 4-Methyl-1 H-indazole
- 2-(2-Nitrophenyl)ethanol
- 2-Cyano-N-ethylacetamide
- Oct-1-yn-3-ol
- N, N-Dimethylpiperidine-4-carboxamide hydrochloride
- Methyl-3-(4-hydroxyphenyl)propionate
- 7,8-Difluoroquinoline
- 1,3-Dicyclohexylthiourea
- 2,2,2-Trimethylacetophenone
- N-(2-Methyl-4-oxopentan-2-yl)acrylamide
- 2-(1H-Pyrrol-1-yl)aniline
- 1,10-Phenanthroline monohydrochloride monohydrate
- 6,7-Dihydro-3H-cyclopenta[4,5]thieno[2,3-d]pyrimidine
- 1-Phenylindoline-2,3-dione
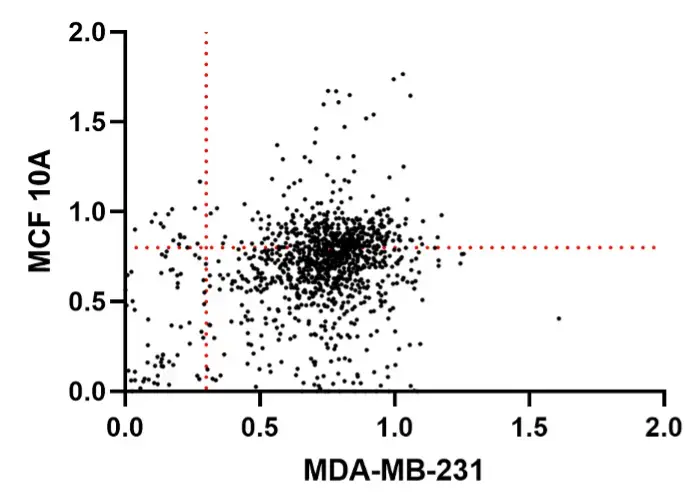
Figure 1. Scatter plot of drug screening data
After conducting another round of standardized drug screening, we performed multiple additional validations on the MCE Novel Known Bioactive Compound Library by subjecting them to high-throughput drug screening. This time, we identified two small-molecule compounds: 6,7-Dihydro-3H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4(5H)-one and 1,10-Phenanthroline monohydrochloride monohydrate.
Figure 2. The structure of 6,7-Dihydro-3H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4(5H)-one
Figure 3. The structure of 1,10-Phenanthroline monohydrochloride monohydrate
Since 6,7-Dihydro-3H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4(5H)-one (12F4) has demonstrated stable anti-tumor effects, our team conducted cellular functional validation using this drug. The result showed that 12F4 had relatively low toxicity in normal breast epithelial cells, while it exhibited stronger toxicity in breast cancer epithelial cell lines.
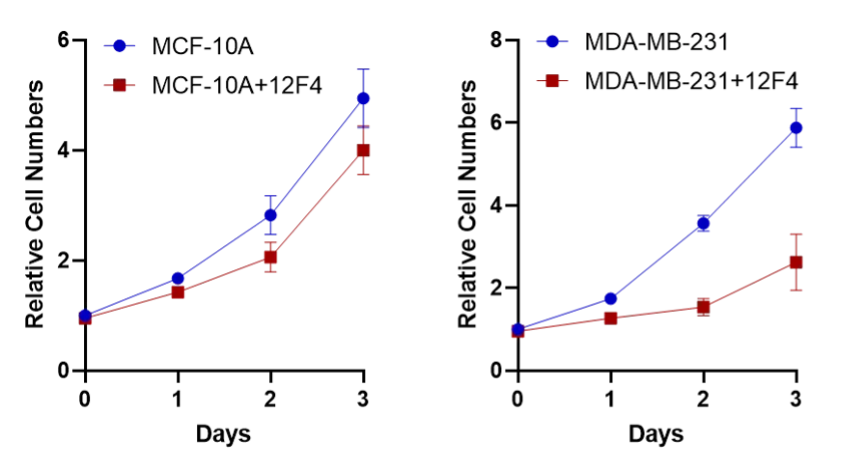
Figure 4. The cell growth curve of 12F4
Virtual Screening
Through virtual screening, we can further validate the results of the wet experiment. We performed molecular docking between the drug small molecules and four target proteins in the drug library and finally selected some drugs with high comprehensive scores. More details about the four target proteins are on the modeling page. The overlap between the virtual screening results and the experimental results is as follows.
- 1,10-Phenanthroline monohydrochloride monohydrate
- 6,7-Dihydro-3H-cyclopenta[4,5]thieno[2,3-d]pyrimidine
- 2,2,2-Trimethylacetophenone N-(2-Methyl-4-oxopentan-2-yl)acrylamide
- 7,8-Difluoroquinoline Propylbenzenesulfonate
- 3-Morpholin-4-yl-propionic acid hydrochloride
- 1-(4-Methylphenyl)piperazine 4-Methyl-1 H-indazole
The following are the docking results of the two screened drugs and our four target proteins. Most of the drugs that were screened through experiments obtained high-scoring functions in molecular docking, which confirmed the scientific validity of our experimental screening results.
Figure 5. mTOR as docked with 1,10-Phenanthroline monohydrochloride monohydrate
Figure 6. mTOR as docked with 6,7-Dihydro-3H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4(5H)-one
Opening multiple molecular visualizations simultaneously may cause severe browser lag.
Do you wish to continue?
Figure 7. CDK4/6 as docked with 1,10-Phenanthroline monohydrochloride monohydrate
Opening multiple molecular visualizations simultaneously may cause severe browser lag.
Do you wish to continue?
Figure 8. CDK4/6 as docked with 1,10-Phenanthroline monohydrochloride monohydrate
Opening multiple molecular visualizations simultaneously may cause severe browser lag.
Do you wish to continue?
Figure 9. EGFR as docked with 1,10-Phenanthroline monohydrochloride monohydrate
Opening multiple molecular visualizations simultaneously may cause severe browser lag.
Do you wish to continue?
Figure 10. EGFR as docked with 6,7-Dihydro-3H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4(5H)-one
Opening multiple molecular visualizations simultaneously may cause severe browser lag.
Do you wish to continue?
Figure 11. BRCA as docked with 1,10-Phenanthroline monohydrochloride monohydrate
Opening multiple molecular visualizations simultaneously may cause severe browser lag.
Do you wish to continue?
Figure 12. BRCA as docked with 6,7-Dihydro-3H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4(5H)-one
Discovery of Modifiable Structures
By analyzing the properties of the molecules themselves and how they bind to each other, we can improve the structure of the drug to better bind to the target protein.
We selected the drug that scored highly in molecular docking, which was also one of the drugs that we screened experimentally. We analyzed its structure and binding to the target protein, and recommended improvements to the structure of the drug based on this information. The improved drugs achieved higher scores after molecular docking analysis. More details are available on our Model page.
Through our research, we found no previous reports on the anti-cancer properties of these two compounds, suggesting that they may hold potential for further development in the future and could offer new therapeutic targets for breast cancer treatment. We utilized molecular docking technology to screen drugs from a drug library and validated the scientific accuracy of the wet lab screening results. Both of the final compounds identified were included in the molecular docking results, with high docking scores, confirming the effectiveness of our drug screening experiment and the ScanCer tool.
Summary
ScanCer is an ATP-FRET sensor for high-throughput breast cancer medicine screening. Using the biosensor that we have developed, we discovered the anti-cancer properties of two small molecule compounds.
It is evident that as a high school student team, there are still many areas that require improvement. However, we put in great effort to turn our ideas into reality, implement them, and continuously enhance their overall performance.
References

